│
└───Data
│ │
│ └───Input
│ │ │
│ │ └───Chronology_setting
│ │ │ │
│ │ │ └───Bchron_crash
│ │ │ │
│ │ │ └───Chron_control_point_types
│ │ │ │
│ │ │ └───Percentage_radiocarbon
│ │ │
│ │ └───Depositional_environment
│ │ │ │
│ │ │ └───Neotoma
│ │ │ │
│ │ │ └───Other
│ │ │
│ │ └───Eco_group
│ │ │
│ │ └───Harmonisation_tables
│ │ │
│ │ └───Neotoma_download
│ │ │
│ │ └───Potential_duplicates
│ │ │
│ │ └───Other
│ │ │
│ │ └───Regional_age_limits
│ │
│ └───Personal_database_storage
│ │
│ └───Processed
│ │
│ └───Chronology
│ │ │
│ │ └───Chron_tables_prepared
│ │ │
│ │ └───Models_full
│ │ │
│ │ └───Predicted_ages
│ │ │
│ │ └───Temporary_output
│ │
│ └───Data_filtered
│ │
│ └───Data_harmonised
│ │
│ └───Data_merged
│ │
│ └───Data_with_chronologies
│ │
│ └───Neotoma_processed
│ │ │
│ │ └───Neotoma_chron_control
│ │ │
│ │ └───Neotoma_dep_env
│ │ │
│ │ └───Neotoma_meta
│ │
│ └───Other
│
└───Outputs
│
└───Data
│
└───Figures
│ │
│ └───Chronology
│ │
│ └───Pollen_diagrams
│
└───Tables
│
└───Meta_and_referencesA step-by-step guide to data processing with FOSSILPOL
Summary
The FOSSILPOL Workflow is structured in a modular manner, where all steps are organised sequentially and guided by one main configuration file (Config file) where all criteria and setup configurations are pre-defined by the user.

Figure 1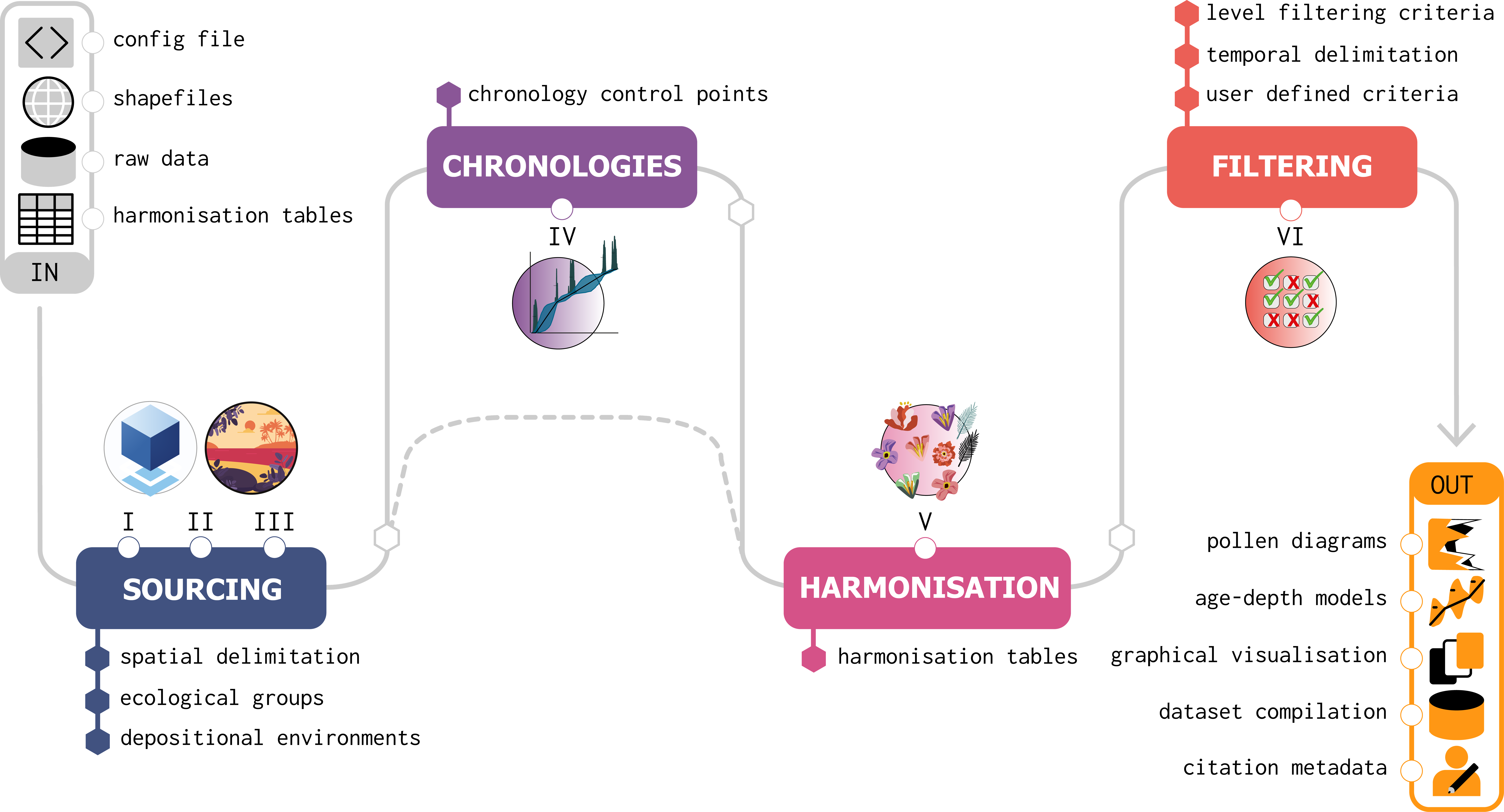
Data input
The FOSSILPOL workflow is set up in a way that data from Neotoma Paleoecological Database (“Neotoma” hereafter) are the primary data input. However, other data sources can also be used in parallel by using our predefined format (Fig. 2). The user thus has the flexibility to source data from either Neotoma or from another data source as long as our predefined format file is used (see other data sourcing).
Three additional data inputs are required for the initial set-up of the Workflow:
Configuration file (
00_Config_file.R) - this contains all the user-selected settings which will be applied throughout the Workflow. These range from technical settings (e.g. location of the data storage) to specific requirements (e.g. filtering criteria) for records to be included. An overview of where the config critera are used through the Workflow is summaried in (Fig. 2).Geographical shapefiles - the workflow is internally set-up in a way that data are processed by geographical regions and shapefiles are used to assign relevant geographical information to the records to process. First, the Workflow is conceptualised for a global project, so the general structure of data processing is done per continent (i.e.
region= “continent”), but the user can use any other delineation of interest. The Workflow comes with a default shapefile roughly delimiting continents, but it can be adjusted or replaced as per project needs. Second, the taxonomic harmonisation of records is structured by harmonisation regions provided byharmonisation region shapefile. By default, this shapefile is a copy of the continental shapefile, but as harmonisation tables are region-specific (see next data input item) this shapefile needs to be adjusted to represent the geographical delimitation of the harmonisation regions used. Finally, if the user is interested in other biogeographic, climatic, or ecological units of interest to be linked to each record (e.g. ecozones, biome type, climate zones), then additional shapefiles (or TIF files) can be added to the workflow (see details here).Harmonisation tables - in each project, one harmonisation table must be provided per harmonisation region (delimited by the corresponding harmonisation region shapefile, see above). A harmonisation table always comes with two columns: i)
original taxa(original taxa) with taxonomic names originally present in Neotoma and/or other data source in the project, and ii)level_1(harmonised taxa) with the standardised taxonomic names. The Workflow will detect if a harmonisation table has been provided by the user, or otherwise create a new table with all detected raw taxa names for each harmonisation region. The latter can consequently serve as a template for harmonisation in thelevel_1column (see details here).
Figure 2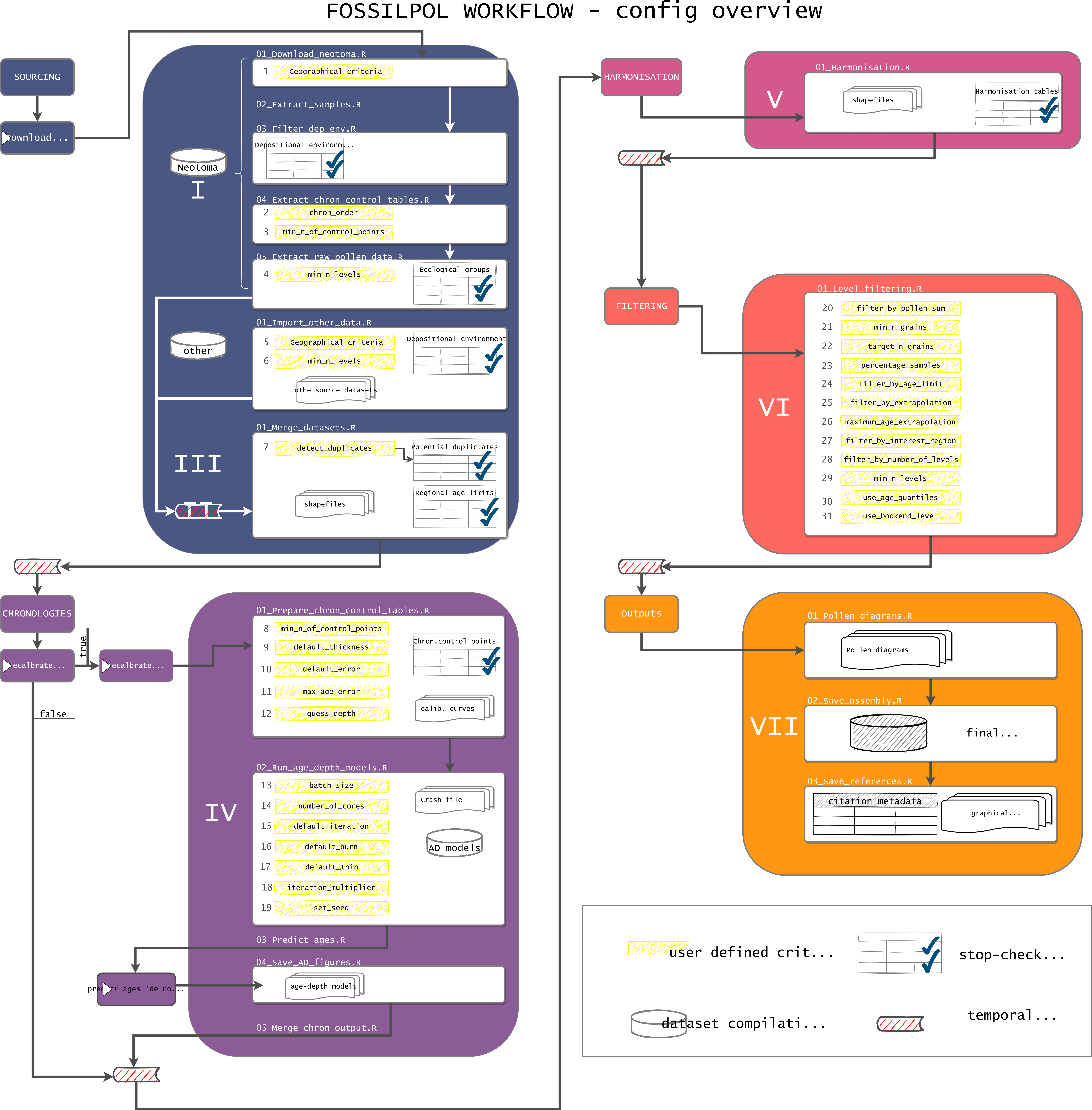
Data storage
The Workflow will produce several files including temporary output files, stop-check tables, and final outputs (data assembly, figures, etc.):
Temporary output files: the Workflow is set up in a way that temporary (in-progress) data files are saved at various stages of the Workflow. Each file will contain the date of creation for easier organisation. When run multiple times, the Workflow will automatically detect if there are any changes in a selected file and only overwrite it, if an updated file is produced (this is powered by {RUtilpol} package). This also means that the user does not have to re-run the whole Workflow but can re-run only specific parts. As the total size of the files can become substantial, the user can specify if all files should be stored within the project folder (default) or in another directory (specified by using the
data_storage_pathin the Config file). With such specification, and after running the00_Config_file.Rscript, there will be an additional folder structure created (see Code block 3).stop-checks CSV tables: while running the Workflow, there will be several times that a user will be asked to check and, where necessary, adjust the produced CSV tables to subsequently continue with the Workflow (i.e. re-run script). This is done to oblige the user to check the produced intermediate results before continuing. For example, at a certain point, the Workflow will produce a list of all ecological groups detected within the dataset compilation obtained from Neotoma. A user then has to edit the mentioned CSV table and specify, which ecological groups should be kept (
include=TRUE) and which should be filtered out (include=FALSE). Note that there are several stop-checks throughout the Workflow (see overview in Fig. 2).Workflow output (
Outputs/, see Section VII for more information):- a ready-to-use, taxonomically harmonised and temporally standardised compilation of fossil pollen data, ready for the analytical stage (rds format)
- plots of modelled age-depth curves for each record (pdf format)
- pollen diagram of each record (pdf format)
- metadata table relaying the main data contributor, contact information, and corresponding publications for citation purposes of the used datasets (PDF).
- reproducibility bundle, a zip file with contains all important sections for a reproducibility of the whole project.
- overview figures of the spatial and temporal distribution of the dataset compilation, namely a map and a graph of the record lengths, respectively (PDF).
Code block 3
Data processing
Here we focus on the scripts within the R/01_Data_processing folder representing all steps needed for data processing (from obtaining data to the final dataset compilation), organised in the following Sections:
Data sourcing:
/01_Neotoma_source/- retrieve and process data from NeotomaData sourcing:
/02_Other_source/- process data from other data source (optional)Initial data processing:
/03_Merging_and_geographic_delineation/- merge data sources, filter out duplicates, and assign values based on geographical locationChronologies:
/04_Chronologies/- prepare chronology control tables, calculate age-depth models, and predict ages for levelsHarmonisation:
/05_Harmonisation/- prepare all harmonisation tables and harmonise pollen taxa (morphotypes)Data filtering:
/06_Main_filtering/- filter out levels and records based on user-defined criteriaOutputs:
/07_Outputs/- save the final output including dataset compilation, pollen diagrams, metadata information, graphical summary snd reproducibility bundle
I. Data sourcing: /01_Neotoma_source/
Run_01_01.R- run all scripts within this folder01_Download_neotoma.R- download the pollen data from the Neotoma database02_Extract_samples.R- create a table from Neotoma’s downloaded lists and download author information03_Filter_dep_env.R- get depositional environment data and filter out records based on the user preferences04_Extract_chron_control_tables.R- get chronologies, including the preferred table with chronology control points05_Extract_raw_pollen_data.R- extract the raw pollen counts from Neotoma and filter by user-selected ecological groups.
01_Download_neotoma.R
All pollen records are downloaded from Neotoma based on the geographical criteria (spatial extent, Fig. 2 - config criteria 1) and the selected data type, in this case: "pollen". Note that a more complex spatial extent, like a polygon, can be used with the loc argument in RFossilpol::proc_neo_get_all_neotoma_datasets()(see usage of loc in neotoma2 example here).
02_Extract_samples.R
Each record is processed using a unique dataset ID (dataset_id) with metadata information extracted. Metadata includes information about the name of the record, geographical information, and the authors and publication DOI connected to the dataset. The authors and their link to the dataset are saved into a separate Author-Dataset database created specifically for each project. It allows easy extraction of authors and DOI for the final dataset compilation produced by the Workflow.
03_Filter_dep_env.R
Depositional information from each record gives information about the environments where a record was extracted. Based on the research question, there could be a preference for certain environments (e.g. terrestrial vs. marine). Currently in Neotoma, the data about depositional environments are organised in a hierarchical structure (e.g. “Pond” is nested in “Natural Lake”, which is nested in “Lacustrine”), in which the maximum number of nested layers is five. At the lowest hierarchical level, there are currently over 50 different categories of depositional environments (for fossil pollen records). Based on the selected records, the Workflow will produce a list of all depositional environments (and their hierarchical position) present in the user´s data selection. The user is then requested to define the environments of choice (this is a stop-check point, Fig. 2). Note that excluding depositional environments with the higher hierarchical position will not automatically exclude all depositional environments nested in it.
04_Extract_chron_control_tables.R
Chronology data for each record is available in a table that contains information about chronology control points used to construct an age-depth model. Some records can have multiple chronology tables as some records have been used for several projects or recalibrated (updated) by data stewards. These tables are numbered according to the order in which they were created and uploaded. Each chronology comes with the age-unit of the age-depth model output (e.g. “Radiocarbon years BP”, “Calibrated radiocarbon years BP”) and the temporal range of the record (youngest and oldest age). The chronologies in “Radiocarbon years BP” are often older chronologies as it is now common practice to recalibrate radiocarbon-dated material and produce chronologies expressed in “Calibrated radiocarbon years BP”. Note: The chronologies in “Calibrated radiocarbon years BP” still come with chronology table(s) containing the uncalibrated radiocarbon ages and need to be calibrated by the user if a new age-depth model is desired. The Workflow automatically selects one table per record based on the order defined by chron_order in the Config file (Fig. 2 - config criteria 2). Note: if more tables have the same age-unit type (e.g. Calibrated radiocarbon years BP), the Workflow will opt for the more recent table. The user can specify their preference for certain age-unit types in the Config file. In addition, only records which have at least a certain number of control points (defined by min_n_of_control_points in the Config file, Fig. 2 - config criteria 3) will be subsequently used.
05_Extract_raw_pollen_data.R
Each level of each record comes with additional information: a) unique sample ID (sample_id), b) information about depth (and estimated age later), and c) pollen counts for each taxon present in that level. The information about levels is split into two different tables (first with depth and ages, and second with pollen counts) linked together by sample ID (sample_id).
The Workflow will only keep records with a minimal number of levels as defined in the Config file (min_n_levels, Fig. 2 - config criteria 4). The minimum number of levels is by default selected as three but the user can change this setting.
In the case of data sourced from Neotoma, each pollen taxon has information about the ecological group (e.g. palms, mangroves, etc). Based on the selected records, the Workflow will produce a full list of all ecological groups after which the user is requested to define which ecological groups to include (a stop-check point, Fig. 2, see explanation of abbreviation in Table 1)
Table 1. Ecological groups assigned to pollen taxa as defined in Neotoma
| ABBREVIATION | ECOLOGICAL GROUP |
|---|---|
| ACRI | Acritarchs |
| ANAC | Anachronic |
| ALGA | Algae (e.g. Botryococcus) |
| AQB | Aquatics (e.g. Sphagnum) |
| AQVP | Aquatic vascular plants (e.g. Isoetes) |
| BIOM | Biometric measurements |
| EMBR | Embryophyta |
| FUNG | Fungi |
| LABO | Lab analyses |
| MAN | Mangroves |
| PALM | Palms |
| PLNT | Plant |
| SEED | Unidentified, but definitely pollen - Spermatophyte rank or clade |
| SUCC | Succulents |
| TRSH | Trees and shrubs |
| UNID | Unknown and Indeterminable |
| UPBR | Upland bryophytes |
| UPHE | Upland herbs |
| VACR | Terrestrial vascular cryptogams |
| VASC | Vascular plants |
II. Data sourcing: /02_Other_source/
Run_01_02.R- run all scripts within this folder01_Import_other_data.R- source other data sources and filter the records in a similar way as Neotoma.
Note on other data sourcing
Our FOSSILPOL Workflow allows the use of other data source in combination with the Neotoma data. Including other data sources is fully optional and can be skipped as indicated by use_other_datasource = TRUE/FALSE in the Config file.
Any data can be used as long as it contains the following required information: a) metadata, b) depositional environment, c) chronology, d) level (age-depth), and e) pollen counts. In order to prepare data for usage, the user needs to download file template specially prepared for this. Each pollen record needs to be stored as a separate file with unique name. We recommend e.g., private_data_(insert_site_name).xlsx. The site name in the filename is crucial as it will be compared against all other pollen records in Neotoma to test for potential duplicates in a later stage of the Workflow. All files must be stored in /Data/Input/Other/ (or specified by dir_files argument, see below).
01_Import_other_data.R
The sourcing of other data sources follows a simple order of actions:
Data files need to be prepared by the user following the template one record per file.
Data are extracted and formatted to be compatible with Neotoma data using the
RFossilpol::import_datasets_from_folder()function, with the following arguments:dir_files- user can specify which folder contains the prepared data (default =Data/Input/Other/suffix- argument to keep track of the source of the data. Default is set to"other", which means that datasets can be easily identified as their name will be(dataset id)_othersource_of_data- will flag the source of each dataset in the compilation in meta-data overview (see section VII). Default is set to"personal_data"data_publicity- will flag the data publicity of each dataset in the compilation in meta-data overview (see section VII - Outputs). Default is set to"restricted"pollen_percentage- pollen counts measured as proportions (e.g., from scanning of pollen diagrams) can be flagged here. Default set toFALSE
Names of data contributors are extracted and added to the Author-Dataset database used in Author-dataset attribution (see section VII - Outputs).
Data are treated in a similar way as data from Neotoma, in terms of filtering by geographical location, number of levels (Fig. 2 - config criteria 5,6), and depositional environments (stop-check point, Fig. 2).
III. Initial data processing: /03_Merging_and_geographic_delineation/
Run_01_03.R- run all scripts within this folder01_Merge_datasets.R- merge data from all sources, filter out duplicates, and assign values based on geographical location
01_Merge_datasets.R
After initial data processing, records from Neotoma and Other sources are merged together.
Detection of duplicates
There is a possibility that some datasets from the other data sources are already in Neotoma. To avoid duplication within the final dataset compilation, the Workflow will compare datasets from both sources and identify potential duplicates. This step is optional but recommended to follow. To do so, the user needs to specify that detect_duplicates == TRUE in the Config file (this is set as default, Fig. 2 - config criteria 7). The Workflow will start a simple subroutine using the function RFossilpol::proc_filter_out_duplicates(). Because comparing all records within each data source to each other is relatively computationally demanding, the function will split the data into several groups using their geographical location (ca. 100 records per group). The user can define the number of groups using the n_subgroups argument. Next, each record from one source is compared to all records from the other source as long as they are within 1 degree radius (the assumption here is that duplicated records will be in a similar location). The user can define the maximum distance using the max_degree_distance argument. Finally, the Workflow will output a list of potential duplicated records (a stop-check point, Fig. 2). For each record-pair, the user must specify, which records should be deleted by writing 1 (deleting the Neotoma) or 2 (deleting the other data source) in the delete column of the created list (leaving 0 will leave both records in).
Additional data preparation
Several more steps take place to create the fully merged dataset compilation before proceeding to the chronology step (does not require any action by user):
All taxon names are transformed into a more computer-friendly format for easier manipulation. The
RFossilpol::proc_clean_count_names()function will first transform special characters to text (e.g.+to_plus_) and then use the {janitor} to transform into “snake_case” style. In addition, the user can specify additional specific changes in names (e.g. based on presence of special characters) by adjusting theuser_name_patternsargument (see example in the script). During this cleaning of taxa names, the Workflow will savetaxa_refrence_tablefor back traceability to Neotoma taxonomy. Thetaxa_reference_tableis a CSV file saved in the same folder as the harmonisation tables (Data/Input/Harmonisation_tables/). More about harmonisation process.Individual levels (sample depths) are sorted by their depth for each record by
RFossilpol::proc_prepare_raw_count_levels(). This includes subroutines, for example, only keeping levels present in all data tables, filtering out levels without pollen data, and taxa which are not present in any level.Spatial information for each record is assigned based on the provided geographical shapefiles. Specifically:
Region information - the shapefile in
Data/Input/Spatial/Regions_shapefilewill assign the regional names for each record (see Data input Section). The user can (and is recommended to) change the spatial delineation of the data by altering or replacing the shapefile.Political delineation (countries) - obtained from GADM database, version 2.8, November 2015.
Harmonisation region - the shapefile in
Data/Input/Spatial/Harmonisation_regions_shapefilewill assign the harmonisation region (to be able to link the corresponding harmonisation table to use; See see Data input Section). The default shapefile in the Workflow is a copy of the Region information shapefile but should be adjusted by the user to correspond to the area covered by the different harmonisation tables.Calibration curves (normal and post-bomb) - depending on the geographical position of the record, a different calibration curve needs to be assigned, as different curves are used for the northern and southern hemispheres, and for terrestrial and marine environments. See more details about calibration curves.
Additional - The user can add any additional spatial delimitation (e.g. ecozones). This will require adding the specific shapefile (or TIF file) in
/Data/Input/Spatial/NAME_OF_FOLDERand adjusting the R code manually (optional_info_to_assign) so that the shapefile is sourced, and its information assigned to each record (see the example in the script).
The Workflow will create a new table with age limitations for each region presented in the data, which needs to be edited by the user (a stop-check point, Fig. 2). For example,
Regional_age_limitstable will have the following values:young_age= youngest age the record must haveold_age= oldest age the record must haveend_of_interest_period= levels beyond this age will be omitted
IV. Chronologies: /04_Chronologies/
Run_01_04.R- run all scripts within this folder01_Prepare_chron_control_tables.R- prepare the chronology tables for age-depth modelling02_Run_age_depth_models.R- create age-depth models with BChron03_Predict_ages.R- estimate the ages of individual levels04_Save_AD_figures.R- save visual output of the age-depth models in pdf format05_Merge_chron_output.R- link age-depth models to corresponding datasets
Note on age-depth modelling
To estimate the age of individual levels based on their depth, an age-depth model needs to be constructed based on the chronology data of the record. An age-depth model will provide age estimates of each individual level and the full age range of the record.
Age-depth modelling can be very computationally heavy and can take a substantial amount of time. Therefore, the Workflow automatically processes several files (rds format):
- Chronology-control-tables:
/Data/Processed/Chronology/Chron_tables_prepared/chron_tables_prepared*.rdscontains all the chronology control tables prepared to be re-calibrated
- Age-depth-models:
/Data/Processed/Chronology/Models_full/*contains one file for each age-depth model
- Predicted-ages:
/Data/Processed/Chronology/Predicted_ages/predicted_ages*.rdscontains the predicted ages using the age-depth models
It may be not preferential to re-run all age-depth models every time the user wants to re-run the Workflow. Therefore, the Workflow offers the option to save the successful age-depth models (Age-depth-models & Predicted-ages) and keep them between individual runs. This can be done in several ways:
recalib_AD_modelsin the Config file can be set toFALSE. This will result in completely omitting the scripts aimed at calculating age-depth models, and therefore this step will be skipped. Note that this will only work if the age-depth models have already been successfully created at least once (Predicted-ages exists).calc_AD_models_denovoin the Config file can be set toFALSE(this is the default). When set toTRUE, the Workflow will not use the Age-depth-models files (successful age-depth models) and will re-calibrate everything “de novo”.predict_ages_denovoin the Config file is set toFALSEas the default. When set toTRUE, the Workflow will use all the age-depth model files but re-assign the ages to each level of all related records (even for records where ages were successfully predicted). This is done, for instance, when the number of levels in a record increased since the last Workflow run and these levels still need to have ages assigned.
IMPORTANT: If you select both calc_AD_models_denovo and predict_ages_denovo as FALSE for your first run of age-depth modelling, you might be asked by the Workflow to temporarily switch both to TRUE in the console (you do not have to change anything in the Config file).
01_Prepare_chron_control_tables.R
Age-depth models are constructed using chronology control points (usually radiocarbon dates) with known depth, estimated age, and associated age uncertainties. Each record can and should ideally have several such points saved in the chronology control table. Each chronology control point has the following properties:
- Depth
- Estimated age
- Error of the estimated age
- Type of the chronology control point
- The calibration curve used (which is needed to convert the raw radiocarbon ages to calendar ages (Note: radiocarbon ages are not true calendar ages!)).
This script will take several steps to prepare all records for age-depth modelling, specifically:
- Create and attach the necessary calibration curves
- Select the preferred chronology control point types
- Prepare chronology tables (include fixing issues with percentage carbon (if necessary))
Calibration curves
Calibration curves are assigned to each control point based on several criteria. If a control point has a type flagged to be calibrated (see next section), a calibration curve is assigned based on the geographical position of the record. Note: only chronology control points of uncalibrated radiocarbon ages need recalibration. FOSSILPOL has a shapefile based on figure 7 in Hogg et al (2020) to correctly assigned IntCal20, SHCal20 and a mixed calibration curve to each record. We follow this recommendation “…the use of (i) IntCal20 for areas north of the ITCZ in June-August (dashed lines in Figure 7) which receive NH air masses all year round, (ii) SHCal20 for areas south of ITCZ in December-February (dotted lines in Figure 7) which see SH air masses all year round, and (iii) a mixed curve for areas between the two seasonal ITCZ positions shown in Figure 7, which receive northern air masses in December-February and southern air masses in June-August.” FOSSILPOL also comes with a mixed curve that was constructed using the {rcarbon} package with a proportion of curve contribution 1:1. See more info on mixed calibration curve use.
All calibration curves have an age limitation, i.e. each curve can be used only for certain ages. Radiocarbon calibration curves currently do not cover ages older than 55 thousand years. For the younger ages, during the last century, there are issues with the Earth’s atmospheric radiocarbon and a different set of calibration curves needs to be used. This is caused by the deployment of nuclear weapons in the ’50s and ’60s, which caused a spike in atmospheric radiocarbon and resulted in radiocarbon measurements from material in the following decades to return often highly negative ages. Therefore, if the control point has a radiocarbon age younger than 200 yr BP, a post-bomb calibration curve is used instead (see further below). As with the normal calibration curves, the geographical location of the record is taken into account to assign the corresponding post-bomb calibration curve.
Modern radiocarbon dates are calibrated by using one of the post-bomb calibration curves (nh_zone_1, nh_zone_2, nh_zone_3, sh_zone_1_2, sh_zone_3), following Hua et al., 2013 and link. The Workflow will automatically assign the corresponding curve based on the geographical location of the record (see function IntCal::copyCalibrationCurve()). If modern radiocarbon dates are detected, the Workflow will then display the detected records to the user and apply the conversion according to the post-bomb curves from the {IntCal} package.
Types of chronology control points
Each control point in the control table has several properties: unique ID, depth, age, error, thickness, and chronology control point type (e.g. radiocarbon, biostratigraphic, annual laminations, tephra). Each type of chronology control point has different age uncertainties. For instance, many older records relied on indirect dating techniques based on biostratigraphic layers, similar geological levels from other records (e.g. a volcanic event), and pollen-based levels (e.g. the appearance of a key taxon), among others and can have large age uncertainties into thousands of years. Neotoma has over 50 different chronology controls points that fall within the categories of geochronological (e.g. lead-210, radiocarbon, uranium-series), relative time scale (e.g. MIS5e, Heinrich Stadial 1, Late Wisconsin event), stratigraphic (e.g. biostratigraphic events such as the introduction of anthropogenic taxa), cultural (e.g. European Settlement Horizon), other absolute dating methods (e.g. annual laminations, collection date), and other dating methods (e.g. extrapolated or guesses). Only the chronology control points in uncalibrated radiocarbon ages require recalibration with the calibration curves as most, if not all, other control points will be in calendar ages and no recalibration should be implemented.
A user has the option to select which control point types should be accepted “as-is” and which should be calibrated (a stop-check point, Fig. 2). The Workflow will automatically produce a list of all detected control points from all selected records, which includes columns called include and calibrate. In the first, the user should indicate if the chronology control point should be included. In latter, the user should indicate if the point should be recalibrated using the calibration curves, here uncalibrated radiocarbon ages.
Chronology table preparation
The chronology control tables will need to undergo a number of user-defined adjustments:
filtering out unwanted control point types selected by user (defined by stop-check point, see above)
filtering out records that do not fulfil the minimal number of control points (defined by
min_n_of_control_pointsin the Config file, default =2, Fig. 2 - config criteria 8). The valuemin_n_of_control_pointswill serve as a criterion for the prepared chronology control tables, where records that do not fulfil such requirements will be filtered out. Note that there is a trade-off between accepting only the tables with a high number of control points per time interval (more robust age-depth model) and the number of records that will be able to fulfil strict criteria.fixing instances of missing values. Defined in the Config file, the values
default_thickness(defined in the Config file, default =1, Fig. 2 - config criteria 9) anddefault_error(default =100, Fig. 2 - config criteria 10) will replace missing values (NA) for thickness and error, specifically.filtering out control points with an error that is considered too big. Any control point with an error bigger than
max_age_error(defined in the Config file; default =3000yr, Fig. 2 - config criteria 11) will be filtered out.removing control points that are duplicated in terms of age and/or depth.
in several cases, the chronology control point from the core-top has a type specified as
guess. A user can specify that the typeguessis only acceptable to a certain depth using theguess_depthvariable in the Config file (default is10cm, Fig. 2 - config criteria 12)
In addition, the number and distribution of such control points can give a good indicator of the temporal uncertainty around levels’ ages (Giesecke et al. 2014, Flantua et al. 2016). For example, a record with few chronology control points within the focus time period will have large uncertainties of predicted ages. Hence, the information of the quality of chronologies, i.e. taking into account the types and levels of chronology control points, can be a criterion used in the selection of records.
Percentage carbon
There are three ways by which post-bomb radiocarbon dates are reported, namely by 1) modern radiocarbon dates (<0 F14C yr); 2) percent modern carbon (pMC, normalised to 100%); and 3) fraction radiocarbon (F14C, normalised to 1; Reimer et al. 2004). Currently, there is no direct way to know from Neotoma whether the dates are in pMC or F14C. Even if the cases are likely to be few, such dates need to be checked to avoid incorrect calculations.
The strongest rule of thumb is that normal radiocarbon dates and errors should always be integer (no decimals) and are uploaded so by the data stewards to Neotoma. The second rule of thumb is that pMC control points are characterised by an age value from around 100 with a decimal place. Thus, the Workflow will automatically export suspicious records and the user must specify which control points should be back-transformed (a stop-check point, (Fig. 2).). For those selected by the user to be back-transformed (include == TRUE), the Workflow will first convert the pMC values to normal post-bomb radiocarbon ages (negative 14C ages) by using the IntCal::pMC.age() function, after which normal post-bomb calibration curves are used to calibrate the values to final calendar ages (see above).
Although F14C has been recommended by experts for the reporting of post-bomb samples (Reimer et al. 2004, in practice, the bulk of the reporting is done as modern radiocarbon dates followed by pMC to a much lesser extent. The Workflow currently does not deal with F14C as no such cases have been detected to date. As this is not consistent with the rest of the data, back-transformation could be done when working with other data sources, and the {IntCal} package can be used to do so.
02_Run_age_depth_models.R
Individual age-depth models are estimated using the {Bchron} package, which estimates the Bayesian probabilistic trajectory of the age-depth model curve (non-parametric chronology model to age-depth data according to the Compound Poisson-Gamma model). Therefore, it is suitable for various combinations of control point types, outliers, and age reversals.
If there are many ages at close or similar depths (e.g. annual laminations), initialisation problems may occur and the Bchron could fail to converge the age-depth model. In such a scenario, the thickness of duplicated chronology control points is automatically increased by 0.01 (see artificialThickness argument in Bchron::Bchronology() function).
Multi-core computation
Because creating age-depth models for multiple records can be computationally demanding, the Workflow uses multi-core (parallel) computation. The Workflow automatically detects the number of cores for the machine on which the code is running (this can be adjusted by specifying the number in number_of_cores in the Config file, Fig. 2 - config criteria 14). Several age-depth models are then created at the same time. This is done by splitting the records into batches, with each batch containing a certain number of records (see batch size in Config file, Fig. 2 - config criteria 13; by default it is based on the number_of_cores). If number_of_cores is selected (or detected) as 1, the Workflow will not use the batch approach (as obsolete) and estimate the age-depth models one-by-one (see below).
Note that there is a possibility that the age-depth model estimation will crash (freeze) for unforeseen reasons. Therefore, the Workflow is structured in a way that if an estimation of the whole batch crashes, the Workflow will skip that batch and continue with other batches. The user can specify how long the machine should wait before skipping the batch with the time_per_record argument in the RFossilpol::chron_recalibrate_ad_models() function.
For each batch, the Workflow will try to estimate age-depth models three times. The user can specify the number of attempts by changing the batch_attempts in the RFossilpol::chron_recalibrate_ad_models() function. If the age-depth modelling is stopped in the process, the Workflow will automatically use the previously successful batches, which are saved automatically. This should help when the user needs to stop the age-depth modelling and resume it at a later time.
Next, the Workflow continues to another subroutine where age-depth models for records from failed batches are estimated one by one. Similar to batch estimation, the age-depth model estimation is tried three times for each crashed record until successful at least once. On some rare occasions, a record could cause R to freeze completely and prevents it from skipping to another record. In that case, the dataset ID of the dataset that caused the crash will be automatically written in the Crash file (found in /Data/Input/Chronology_setting/Bchron_crash/) and omitted from the future run of age-depth models. The user is recommended to do a detailed check of the specific dataset, e.g. the chronology control table for possible inconsistencies or other flaws that could be causing {Bchron} to fail to produce an age-depth model.
Iterations
In the Config file, the number of iterations is set to 50,000 by default, discarding the first 10,000 iterations (burn-in) and keeping every iteration beyond the burn-in with a step size of 40 (thinning) (Fig. 2 - config criteria 15,16,17). The user can change the number of iterations by altering the iteration_multiplier in the Config file (Fig. 2 - config criteria 18). This will result in changing the total interactions but keeping the ratios of burn-ins and thinning. Thus 1000 ((50k - 10k) / 40) posterior values are drawn. The default number of iterations should produce a robust estimation but they can be increased by the user if preferred (by increasing the iteration_multiplier). Note that increasing the iteration_multiplier will automatically increase the time that the program will wait for estimation of an age-depth model before skipping it (see time_per_record above).
The user should keep in mind that creating age-depth models for hundreds of records using a high number of iterations is computationally demanding and can take a significant amount of time in the order of tens of hours or several days.
03_Predict_ages.R
With a successful age-depth model, the ages of individual levels are estimated. The Workflow will estimate age and the error estimate for each level, which will encapsulate 95% of all age posterior values (upper and lower boundary). As {Bchron} uses a probabilistic model it is possible to obtain possible ages of each level. To do so, a number of ages (default = 1000, see above) are drawn from the model posterior representing all the possible ages, and a series of quantiles of various values (25, 50, 75, etc.) are then calculated. The 50th quantile (median) is used as the final age of each level, and the 2.5th and 97.5th quantiles as upper and lower boundaries respectively. This results in the age estimate of each level including its error estimates. The whole matrix levels by posterior drawn (possible age) is saved as an age uncertainty matrix (age_uncertainty column). This whole process is completely automated and does not require any input from the user.
04_Save_AD_figures.R
The visual representation of all age-depth models is saved as output for visual confirmation of successful model estimation. The files (as PDFs) are stored in the /Outputs/Figures/Chronologies/ folder and split into subfolders defined by region. The properties of the figures (size of figures, font size, etc) can be altered in the Config file (image_width, image_height, image_units, image_dpi, text_size, line_size). They can be used for inspection of the age-depth curves to look for unrealistically large age estimates or error bars, hiatuses, or extreme extrapolations toward the present or the past.
05_Merge_chron_output.R
The successfully predicted ages are linked with all the records from the various sources (Sections I-III). The individual levels of each record are then ordered by the predicted ages and the same order of levels is then applied to the tables with the pollen counts.
V. Harmonisation: /05_Harmonisation/
Run_01_05.R- run all scripts within this folder01_Harmonisation.R- prepare all harmonisation tables and harmonise the raw counts.
Note on harmonisation
The goal of taxonomic harmonisation is to standardise all site-level names to the same pollen morphotypes (set of pollen and spore morphotypes used for all pollen records) and thus reduce the effect of taxonomic uncertainty and nomenclatural complexity (See relevant literature in Appendix 1 of Flantua et al. 2023). For this purpose, a harmonisation table can be created that groups the morphotypes into the highest taxonomic level that is most likely to be identified by most of the pollen analysts.
01_Harmonisation.R
First, the Workflow will check the harmonisation regions present in the data, defined by the shapefile (see Data input and Section III), and confirm that there is one harmonisation table per region (a stop-check point, Fig. 2). If any table is missing (or the Workflow is run for the first time), the Workflow will automatically create a harmonisation table per harmonisation region, with all the raw taxa names from all the records from within that region. Note that we refer here to “taxon names” for simplicity but the identification is often done at the level of family or genus, and are best referred to as morphotypes.
Each harmonisation table is created so that each taxon can have two columns:
taxon_namewhich is the original name of the taxon (morphotype) formatted into a more computer-friendly format (snake_case)level 1, which should be used to merge various taxa (morphotypes) into harmonised taxonomic units, specific for the project.
Note that in order to link the names to the original display in Neotoma, user can use the taxa_reference_table (see name cleaning process).
The user can also define which taxa should be completely removed during the harmonisation process (marked as delete), in case of a taxonomic mistake or a palaeoecological proxy not of interest, e.g. spores). The user can add additional columns (e.g. level_2) and then specify which levels should be included by altering the argument harm_name when using the RFossilpol::harmonise_all_regions() function.
With these tables, each dataset is harmonised so that all taxa that belong to the same harmonised taxa (morphotypes) are summed together in each level - this process is applicable for both count and percentage data. This process includes automatic detection of successful pollen harmonisation by checking the total number of pollen grains before and after harmonisation (it can be turned off by changing the argument pollen_grain_test to FALSE when using the RFossilpol::harmonise_all_regions() function).
VI. Data filtering: /06_Main_filtering/
Run_01_06.R- run all scripts within this folder01_Level_filtering.R- filter out levels and records based on the user-defined criteria predefined in the Config file
Note on data filtering
To obtain a comprehensive dataset compilation of multiple fossil pollen records, to increase its overall quality, and to answer research questions reliably, we recommend the user to further trim down the data selection by filtering out individual levels and/or whole records. All of these filtering criteria can be adjusted by the user in the Config file:
filter_by_pollen_sum(Fig. 2 - config criteria 20) - ifTRUE, the Workflow will use the quantity of counted pollen grains at each level as a factor in determining the quality of the level.filter_by_age_limit(Fig. 2 - config criteria 24) - ifTRUE, the Workflow will filter out records that do not span the user-defined time period (defined byyoung_ageandold_ageinRegional_age_limitstable; see Section III).filter_by_extrapolation(Fig. 2 - config criteria 25) - ifTRUE, the Workflow will filter out levels based on the number of years between their age and the last chronology control point used for age-depth modelling (chron_control_limits).filter_by_interest_region(Fig. 2 - config criteria 27) - ifTRUE, the Workflow will filter out levels that are older thanend_of_interest_period(defined inRegional_age_limitstable; see Section III) to subsequently reduce processing time during follow-up analyses.filter_by_number_of_levels(Fig. 2 - config criteria 28) - ifTRUE, the Workflow will filter out records based on the number of levels.
In addition, two more options can be activated by the user:
use_age_quantiles(Fig. 2 - config criteria 30) - ifTRUE, the Workflow will use the 95th age quantile (uncertainty of the level age) throughout the data filtration process, i.e. the age of the level will be assumed to be anywhere between the upper and lower boundary (see Section IV). This will result in a more stable dataset compilation between the different results of age-depth modelling (as a probabilistic result can output slightly different results each time they are run). However, the final dataset compilation may require some additional filtering before any analyses, as the 95th age quantile can span very long time periods.use_bookend_level(Fig. 2 - config criteria 31) - ifTRUE, the Workflow will leave one additional level beyond the oldest age of the user-defined time period. This will result in a bookend level, which can help to anchor information beyond the period of interest.
01_Level_filtering.R
Pollen count sum
The number of counted pollen grains at each level is an index of data quality. To obtain a reliable representation of the vegetation, researchers often aim to count more than 300 pollen grains (following Moore et al., 1991), but other recommendations may also have been followed (>150; e.g. Djamali & Cilleros, 2020) and will vary with region and by scientific question (Birks & Birks, 1980). For example, to achieve a representative sample of the regional pollen pool, counts in Arctic records may only reach c. 100 grains per level, whereas counts in Mediterranean sites can be as high as 1000 (Birks & Birks, 1980, p. 165), but the main determinant can also be the preference of the pollen analyst. Reasons for low numbers (<100) are often mainly due to time constraints of the data contributor, but can also be natural depositional phenomena causing poor pollen preservation, such as low local pollen production in arctic or alpine environments. Given that statistical inferential power is proportional to sample size, we recommend defining a minimum number of total pollen grains in each level. Subsequently, whole records can be selected on the proportion of levels with a selected minimum number of pollen grains counted per level.
The user can select two different quantities of total pollen grains per level: a) minimum number (min_n_grains, Fig. 2 - config criteria 21) and b) acceptable number (target_n_grains, Fig. 2 - config criteria 22). All levels with total pollen grains below the minimum number will be filtered out. In addition, the whole record will only be accepted if X% (set by percentage_samples; default = 50, Fig. 2 - config criteria 23) of all levels fulfils at least the acceptable number of pollen grains. This filtration criterion will only be used if filter_by_pollen_sum == TRUE.
Age criteria
As projects differ in their temporal focus, only a subset of records will be of interest to the particular project. Therefore, records that do not span a certain age period (from young_age to old_age; specified by the Regional_age_limits table, see Section III) will be filtered out. This filtration criterion will only be used if filter_by_age_limit == TRUE in the Config file.
Level age extrapolation
Extrapolation of age inferences to samples beyond the set of chronology control points is another factor in quality control. Levels older than the oldest chronology control point have no other chronology control point to constrain the age inference, and therefore, have increasingly large uncertainty as the amount of extrapolation increases. To limit the use of levels based on large extrapolations, we recommend selecting a maximum extrapolation age, i.e. levels older than the selected age criterion (e.g. 5000 yr) from the last chronology control point are filtered out.
In order to limit the use of levels based on large extrapolations, the user can select the maximum extrapolation age (maximum_age_extrapolation, Fig. 2 - config criteria 26), i.e. levels older than the selected criterion from the last chronology control point will be filtered out. This filtration criterion will only be used if filter_by_extrapolation == TRUE in the Config file.
Interest period
The individual levels of a record outside of the interest period (end_of_interest_period specified by the Regional_age_limits table) will be filtered out as they do not provide additional information. This filtration criterion will only be used if filter_by_interest_region == TRUE in the Config file.
Number of levels
The total number of levels in a record is an important quality criterion for further use of such a record in a specific analysis. Records might have been sampled at low resolution (e.g. depth intervals > 30 cm) leaving substantial unassessed gaps - and thus time periods - between levels. In addition, records with few levels will likely contribute poorly to studies focused on specific time periods (many non-value levels) and can cause unnecessary outlier values. Therefore, we recommend selecting a minimum number of levels within the time period of interest and use this as an additional criterion to filter out unwanted records. The user can select the minimum number of levels (min_n_levels, Fig. 2 - config criteria 29) that records must have at the end of the filtration subroutine. This filtration criterion will only be used if filter_by_number_of_levels == TRUE in the Config file.
VII. Outputs: /07_Outputs/
Run_01_07.R- run all scripts within this folder01_Pollen_diagrams.R- save pollen diagrams for all records02_Save_assembly.R- save data assembly with selected variables (columns)03_Save_references.R- save reference and metatable
01_Pollen_diagrams.R
Pollen diagrams for all records will be created using the {rioja} package. The harmonised data will be automatically transformed into relative proportion for plotting purposes. The pollen diagrams will be saved in the /Outputs/Figures/Pollen_diagrams/ folder and split into sub-folders defined by Region (e.g., continent).
The RFossilpol::plot_all_pollen_diagrams() function will automatically produce a PDF of the pollen diagram split into several A4 pages and ready to be printed out. The y-axis is, by default, the age of the levels, but it can be altered to depth (by the y_var argument set to "age" or "depth"). The maximum number of taxa per page can be altered by the max_taxa argument (default = 20). In addition, the function will automatically omit very rare taxa. Specifically, the min_n_occur argument will determine the minimum number of occurrences per record each taxon has to have to be plotted.
02_Save_assembly.R
A ready-to-use, taxonomically harmonised and standardised compilation of fossil pollen data (ready for the analytical stage) is produced and saved in the /Outputs/Data/ folder. The file is saved as a tibble (by {tibble} package from tidyverse) in rds format.
The user can select which columns (variables) should be present in the final data compilation by selecting select_final_variables == TRUE in the Config file. Here, the Workflow will interactively (in the R console) ask the user to specify if each variable should be included (Yes/No).
03_Save_references.R
The workflow will save all metadata and citation information needed to create the dataset compilation. All outputs can be found in the /Outputs/Meta_and_references/ folder. The functionRFossilpol::proc_save_references() will do this automatically but it can be specified by the user, i.e. the user can specify what information should be saved using the user_sel_variables argument. By default this information contains:
"meta_table"- a metadata table (data_assembly_meta.csv) contains the following variables by default (note that this list may be altered by changing the final variables byselect_final_variables):- the list of all records in the final dataset compilation
- geographical location
- depositional environment
- assigned continental region
- assigned harmonisation region
- final number of levels
- number of chronology control points used for age-depth modelling
- assigned calibration curve(s)
- age limits of each record of each data source (Neotoma or other data source)
- DOI (from Neotoma).
"author_table"- a reference table (authors_meta.csv) containing information about the datasets used, the main data contributor, and their contact information."affiliation_table"- if an affiliation is provided with the data from sources other than Neotoma, the affiliation table (affiliation_table) is also exported linking affiliations and their authors."graphical_summary"- a PDF (graphical_summary*.pdf) with three panel figures usingRFossilpol::plot_graphical_summary()function (note that additional arguments can be passed to this function, see?RFossilpol::plot_graphical_summaryfor more information):- map - map of geographical location with each record represented as a point
- count of data records- a lollipop plot with the number of records in each geographical group when assigned
- age length - age limits of records, with each record represented by a single line
"reproducibility_bundle"- a zip file (reproducibility_bundle.zip) of the Config file, all stop-check CSV tables, and all shapefiles. The idea is to increase the reproducibility of the user´s workflow where ideally this zip file can be shared when publishing a publication with a project created using the FOSSILPOL Workflow. For reviewing purposes, the zip file can be shared and detailed feedback can be provided before publication.